Input data and parameters
QualiMap command line
| qualimap bamqc -bam /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_06_bamsort/case1.pair1_sorted.bam -nw 400 -hm 3 |
Alignment
| Command line: | "bismark -q --phred33-quals --bowtie2 --un --multicore 12 --output_dir /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_04_bismark --temp_dir /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_04_bismark --genome_folder /home/zhangwei/Genome/hg19_bismark -1 /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_03_adapterremoval/case1.pair1.truncated.gz -2 /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_03_adapterremoval/case1.pair2.truncated.gz" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | no |
| Program: | Bismark (v0.23.0) |
| Analysis date: | Tue Jan 12 17:42:14 CST 2021 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_06_bamsort/case1.pair1_sorted.bam |
Summary
Globals
| Reference size | 3,137,161,264 |
| Number of reads | 5,237,692 |
| Mapped reads | 5,237,692 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 5,237,692 / 100% |
| Mapped reads, first in pair | 2,618,846 / 50% |
| Mapped reads, second in pair | 2,618,846 / 50% |
| Mapped reads, both in pair | 5,237,692 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Read min/max/mean length | 21 / 150 / 149.09 |
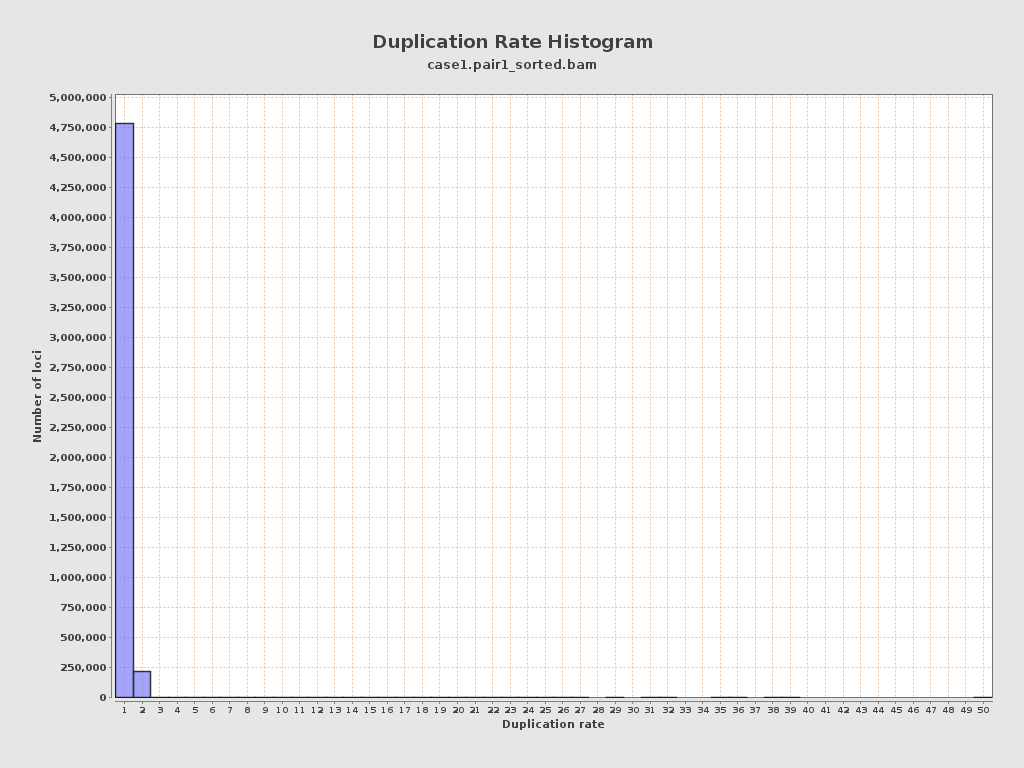
| Duplicated reads (estimated) | 231,706 / 4.42% |
| Duplication rate | 4.42% |
| Clipped reads | 0 / 0% |
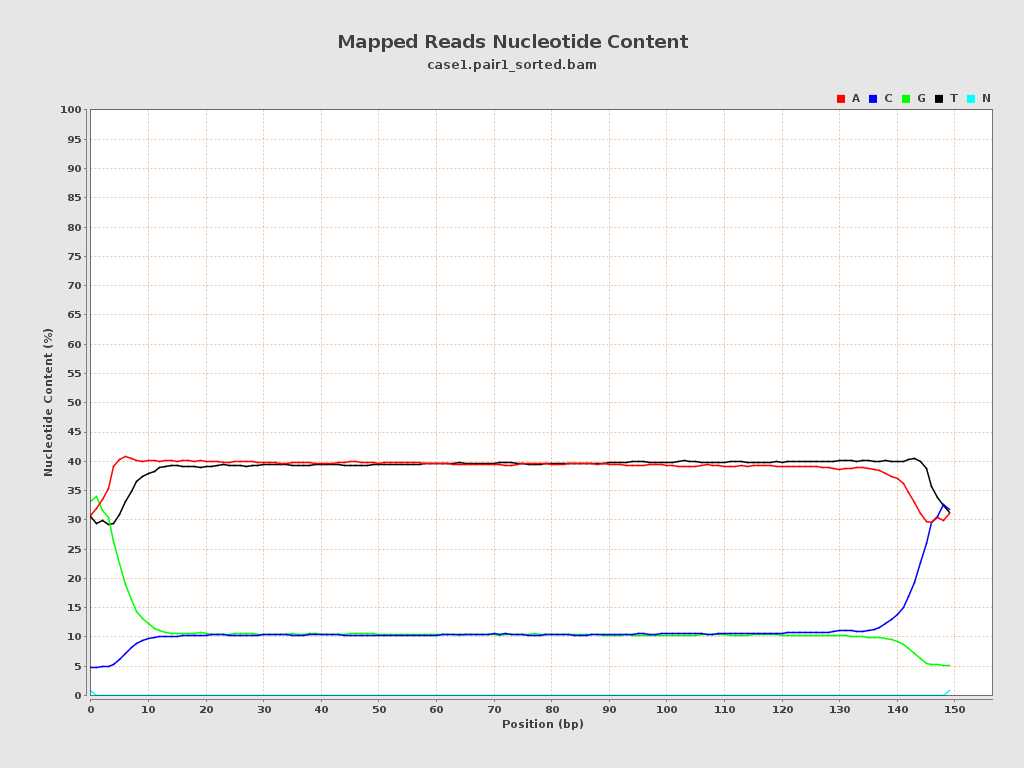
ACGT Content
| Number/percentage of A's | 303,150,109 / 38.95% |
| Number/percentage of C's | 86,063,977 / 11.06% |
| Number/percentage of T's | 303,346,900 / 38.98% |
| Number/percentage of G's | 85,730,601 / 11.02% |
| Number/percentage of N's | 93,413 / 0.01% |
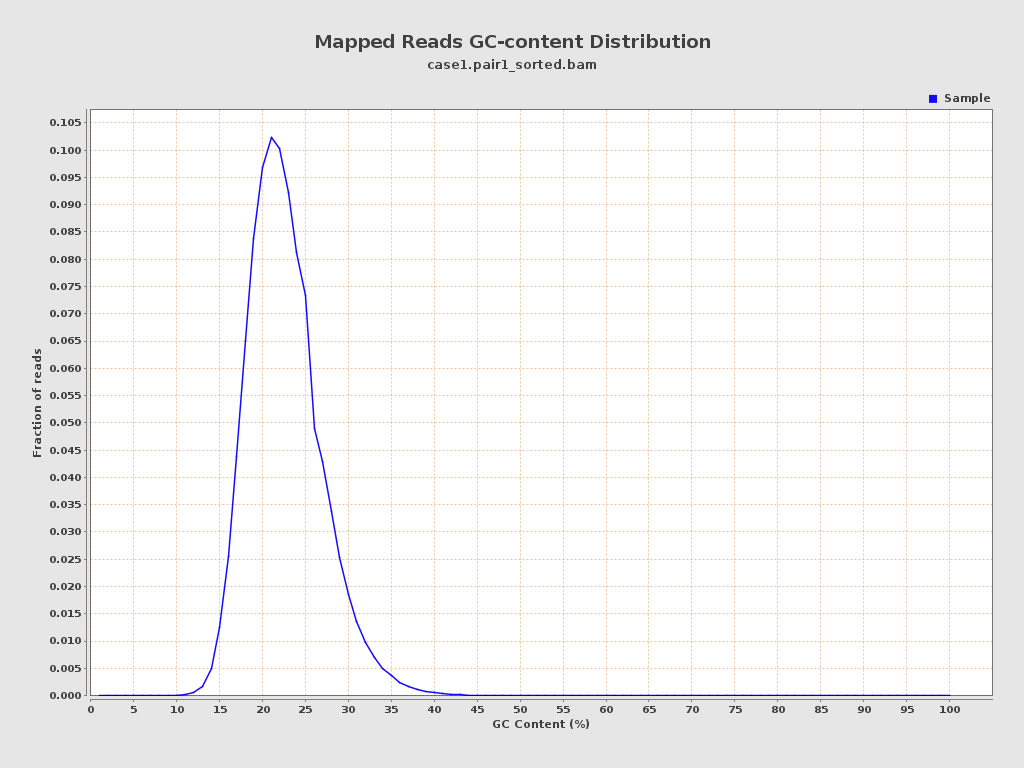
| GC Percentage | 22.07% |
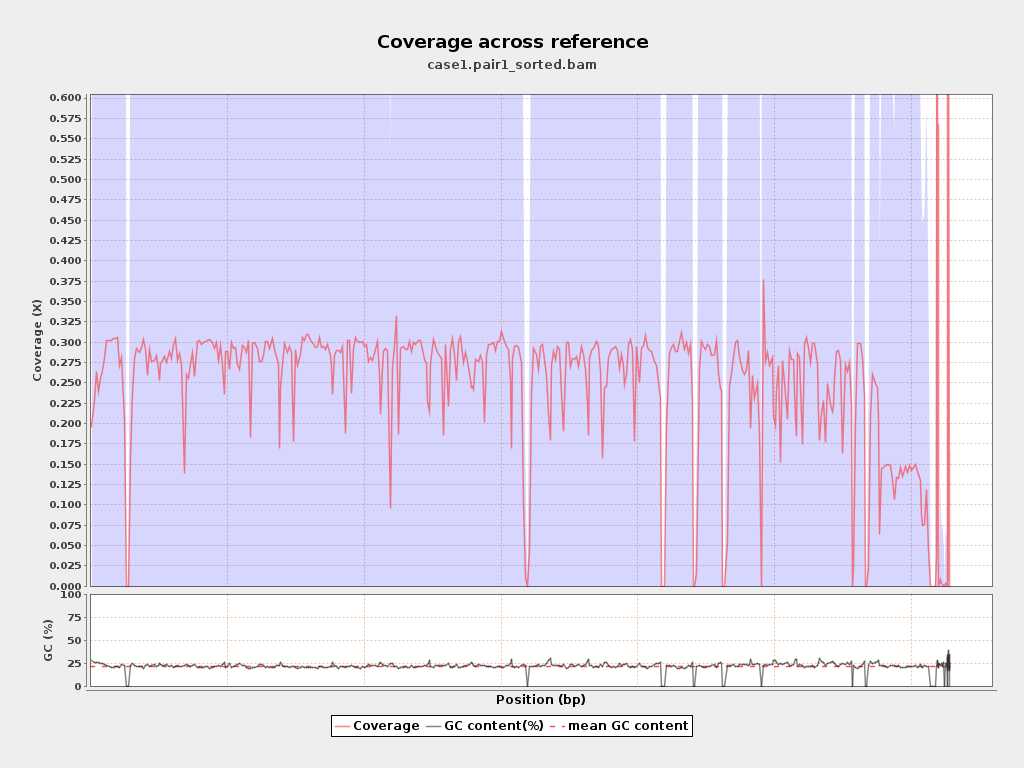
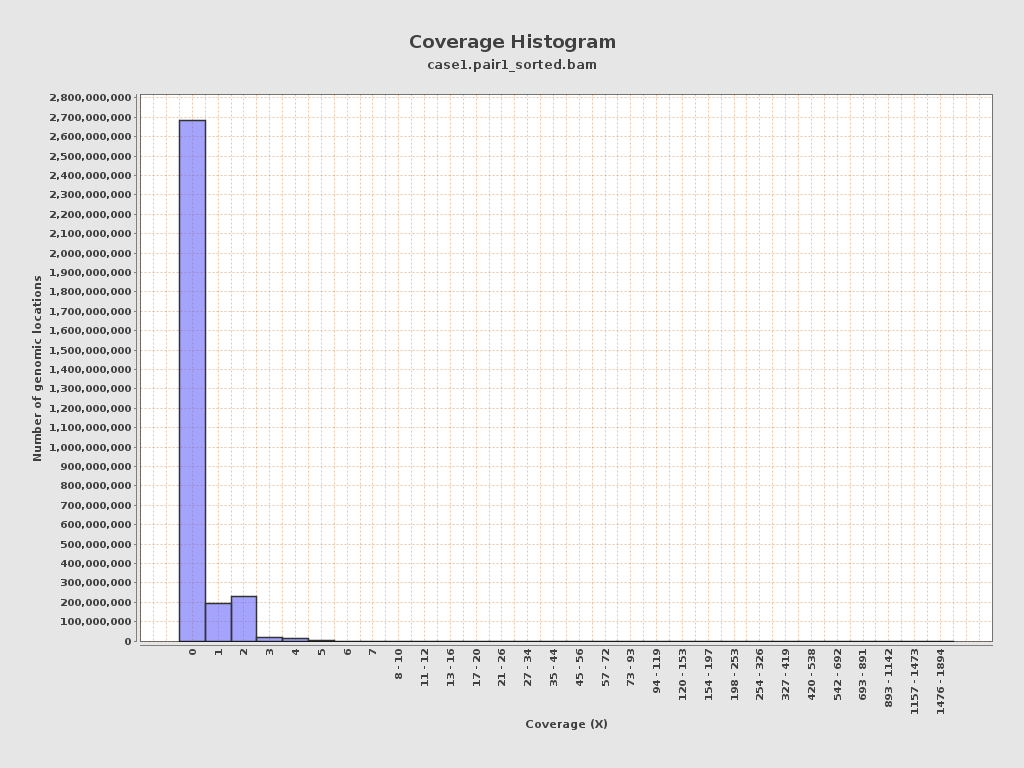
Coverage
| Mean | 0.2485 |
| Standard Deviation | 0.9418 |
Mapping Quality
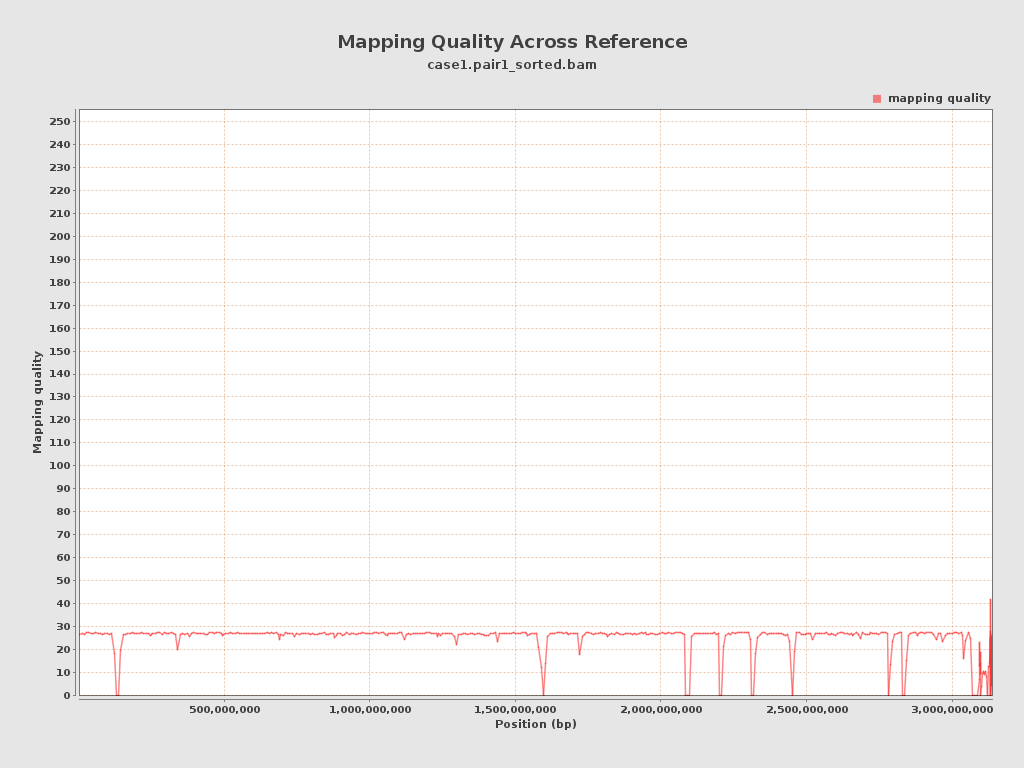
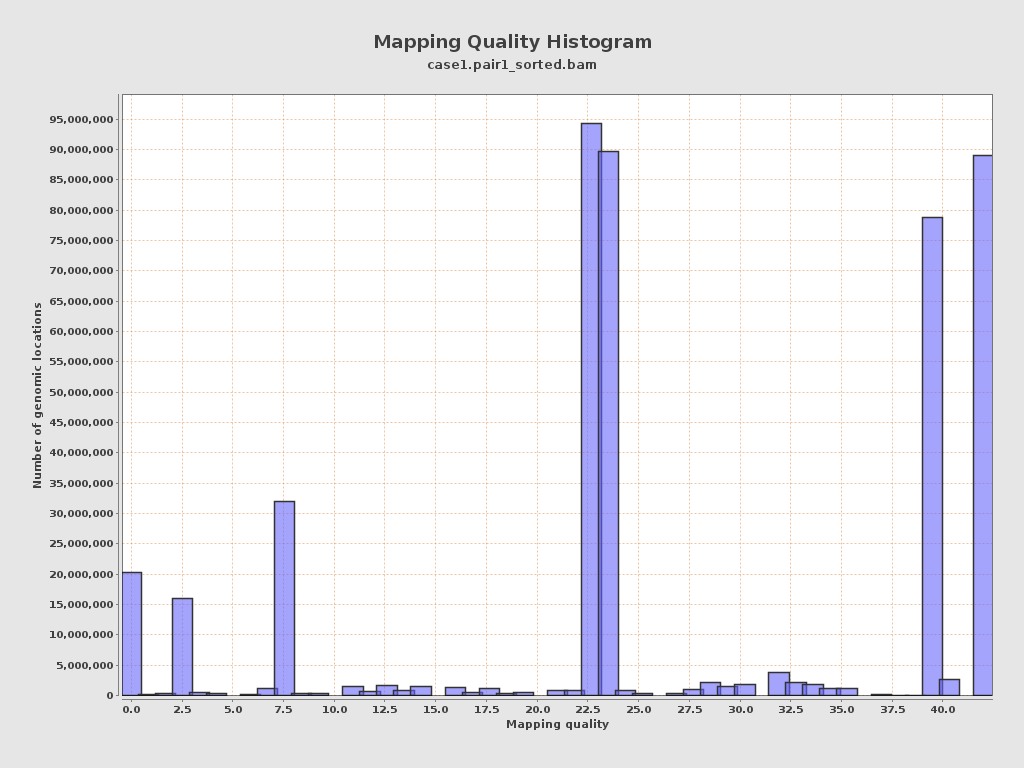
| Mean Mapping Quality | 23.66 |
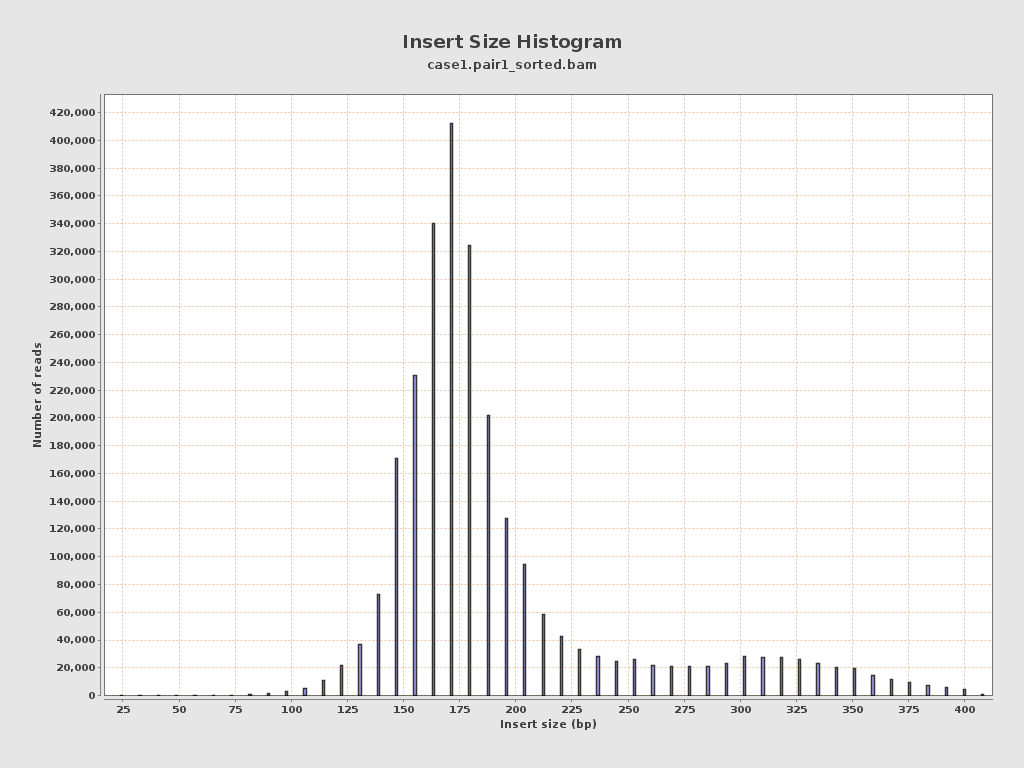
Insert size
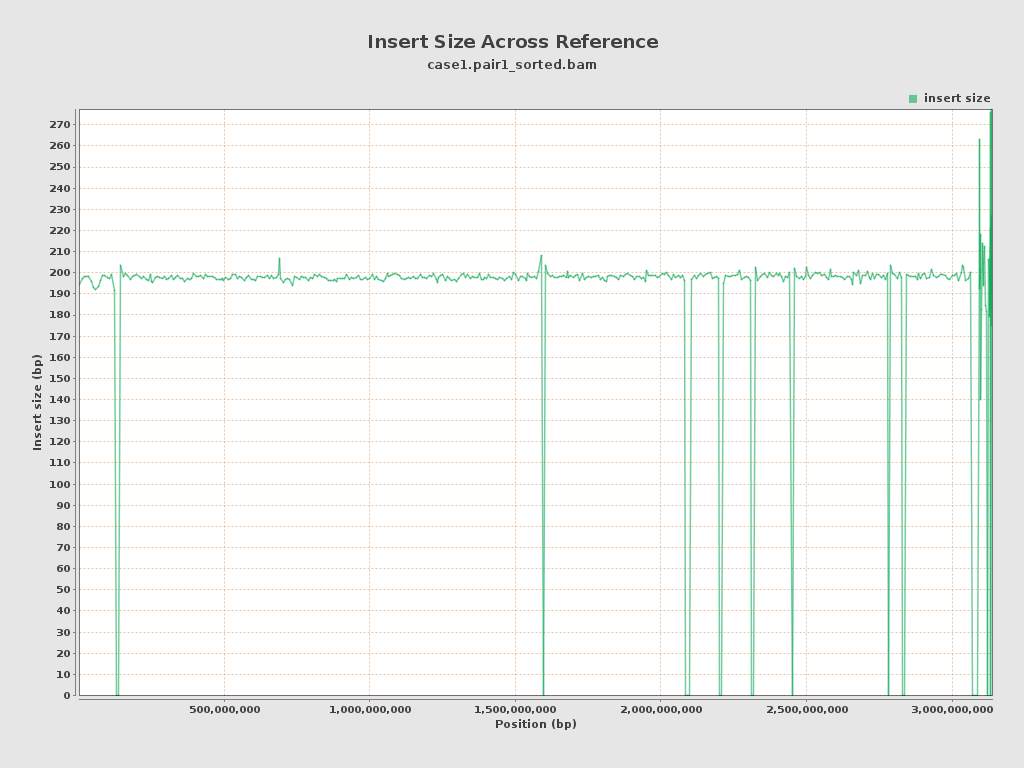
| Mean | 198.61 |
| Standard Deviation | 58.12 |
| P25/Median/P75 | 166 / 180 / 204 |
Mismatches and indels
| General error rate | 20.18% |
| Mismatches | 154,843,251 |
| Insertions | 757,703 |
| Mapped reads with at least one insertion | 14.4% |
| Deletions | 455,014 |
| Mapped reads with at least one deletion | 8.35% |
| Homopolymer indels | 59.36% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| chr1 | 249250621 | 63079022 | 0.2531 | 1.6469 |
| chr2 | 243199373 | 68011573 | 0.2797 | 0.7143 |
| chr3 | 198022430 | 56321635 | 0.2844 | 0.7028 |
| chr4 | 191154276 | 54844170 | 0.2869 | 0.7065 |
| chr5 | 180915260 | 50903218 | 0.2814 | 0.7011 |
| chr6 | 171115067 | 47514029 | 0.2777 | 0.8517 |
| chr7 | 159138663 | 43630040 | 0.2742 | 1.7885 |
| chr8 | 146364022 | 40973990 | 0.2799 | 0.7337 |
| chr9 | 141213431 | 31335761 | 0.2219 | 0.6314 |
| chr10 | 135534747 | 37080081 | 0.2736 | 1.2317 |
| chr11 | 135006516 | 36515552 | 0.2705 | 0.6876 |
| chr12 | 133851895 | 36775475 | 0.2747 | 0.6924 |
| chr13 | 115169878 | 27850649 | 0.2418 | 0.6553 |
| chr14 | 107349540 | 24964450 | 0.2326 | 0.6449 |
| chr15 | 102531392 | 22534280 | 0.2198 | 0.6282 |
| chr16 | 90354753 | 21360309 | 0.2364 | 1.2164 |
| chr17 | 81195210 | 19751299 | 0.2433 | 0.6571 |
| chr18 | 78077248 | 21672839 | 0.2776 | 0.6971 |
| chr19 | 59128983 | 12595180 | 0.213 | 0.7269 |
| chr20 | 63025520 | 16256766 | 0.2579 | 0.6725 |
| chr21 | 48129895 | 10107831 | 0.21 | 0.6231 |
| chr22 | 51304566 | 8285038 | 0.1615 | 0.5427 |
| chrX | 155270560 | 21326524 | 0.1374 | 0.5162 |
| chrY | 59373566 | 2567109 | 0.0432 | 0.4257 |
| chrM | 16571 | 49748 | 3.0021 | 2.2917 |
| chr1_gl000191_random | 106433 | 5672 | 0.0533 | 0.331 |
| chr1_gl000192_random | 547496 | 65176 | 0.119 | 0.4745 |
| chr4_ctg9_hap1 | 590426 | 8095 | 0.0137 | 0.1504 |
| chr4_gl000193_random | 189789 | 107935 | 0.5687 | 1.0747 |
| chr4_gl000194_random | 191469 | 53108 | 0.2774 | 0.7429 |
| chr6_apd_hap1 | 4622290 | 1481 | 0.0003 | 0.0285 |
| chr6_cox_hap2 | 4795371 | 18765 | 0.0039 | 0.0847 |
| chr6_dbb_hap3 | 4610396 | 13415 | 0.0029 | 0.072 |
| chr6_mann_hap4 | 4683263 | 10996 | 0.0023 | 0.0672 |
| chr6_mcf_hap5 | 4833398 | 5531 | 0.0011 | 0.0448 |
| chr6_qbl_hap6 | 4611984 | 10960 | 0.0024 | 0.0626 |
| chr6_ssto_hap7 | 4928567 | 13081 | 0.0027 | 0.0707 |
| chr7_gl000195_random | 182896 | 144186 | 0.7883 | 1.2865 |
| chr8_gl000196_random | 38914 | 300 | 0.0077 | 0.1135 |
| chr8_gl000197_random | 37175 | 304 | 0.0082 | 0.1182 |
| chr9_gl000198_random | 90085 | 26260 | 0.2915 | 0.8459 |
| chr9_gl000199_random | 169874 | 242106 | 1.4252 | 4.8623 |
| chr9_gl000200_random | 187035 | 0 | 0 | 0 |
| chr9_gl000201_random | 36148 | 0 | 0 | 0 |
| chr11_gl000202_random | 40103 | 3589 | 0.0895 | 0.3888 |
| chr17_ctg5_hap1 | 1680828 | 12575 | 0.0075 | 0.1148 |
| chr17_gl000203_random | 37498 | 2704 | 0.0721 | 0.3694 |
| chr17_gl000204_random | 81310 | 8949 | 0.1101 | 0.4656 |
| chr17_gl000205_random | 174588 | 113408 | 0.6496 | 1.1714 |
| chr17_gl000206_random | 41001 | 898 | 0.0219 | 0.1953 |
| chr18_gl000207_random | 4262 | 1190 | 0.2792 | 0.5565 |
| chr19_gl000208_random | 92689 | 68416 | 0.7381 | 1.8245 |
| chr19_gl000209_random | 159169 | 1195 | 0.0075 | 0.1097 |
| chr21_gl000210_random | 27682 | 300 | 0.0108 | 0.1385 |
| chrUn_gl000211 | 166566 | 42077 | 0.2526 | 0.6847 |
| chrUn_gl000212 | 186858 | 42462 | 0.2272 | 0.6394 |
| chrUn_gl000213 | 164239 | 5973 | 0.0364 | 0.2474 |
| chrUn_gl000214 | 137718 | 127903 | 0.9287 | 1.482 |
| chrUn_gl000215 | 172545 | 1498 | 0.0087 | 0.141 |
| chrUn_gl000216 | 172294 | 31332 | 0.1819 | 0.5663 |
| chrUn_gl000217 | 172149 | 15817 | 0.0919 | 0.4019 |
| chrUn_gl000218 | 161147 | 95654 | 0.5936 | 1.0464 |
| chrUn_gl000219 | 179198 | 100114 | 0.5587 | 1.0149 |
| chrUn_gl000220 | 161802 | 1160973 | 7.1753 | 18.5205 |
| chrUn_gl000221 | 155397 | 21713 | 0.1397 | 0.5521 |
| chrUn_gl000222 | 186861 | 13482 | 0.0721 | 0.3335 |
| chrUn_gl000223 | 180455 | 2111 | 0.0117 | 0.1486 |
| chrUn_gl000224 | 179693 | 162710 | 0.9055 | 2.535 |
| chrUn_gl000225 | 211173 | 52923 | 0.2506 | 0.7773 |
| chrUn_gl000226 | 15008 | 220299 | 14.6788 | 42.5859 |
| chrUn_gl000227 | 128374 | 3565 | 0.0278 | 0.2166 |
| chrUn_gl000228 | 129120 | 12864 | 0.0996 | 0.7747 |
| chrUn_gl000229 | 19913 | 7172 | 0.3602 | 0.8901 |
| chrUn_gl000230 | 43691 | 10993 | 0.2516 | 0.7841 |
| chrUn_gl000231 | 27386 | 10968 | 0.4005 | 0.9214 |
| chrUn_gl000232 | 40652 | 28366 | 0.6978 | 1.1725 |
| chrUn_gl000233 | 45941 | 9527 | 0.2074 | 0.5932 |
| chrUn_gl000234 | 40531 | 26200 | 0.6464 | 1.0674 |
| chrUn_gl000235 | 34474 | 18165 | 0.5269 | 1.0015 |
| chrUn_gl000236 | 41934 | 1500 | 0.0358 | 0.2701 |
| chrUn_gl000237 | 45867 | 8384 | 0.1828 | 0.5742 |
| chrUn_gl000238 | 39939 | 1196 | 0.0299 | 0.2121 |
| chrUn_gl000239 | 33824 | 2398 | 0.0709 | 0.3757 |
| chrUn_gl000240 | 41933 | 10142 | 0.2419 | 0.7259 |
| chrUn_gl000241 | 42152 | 33193 | 0.7875 | 1.2854 |
| chrUn_gl000242 | 43523 | 300 | 0.0069 | 0.0889 |
| chrUn_gl000243 | 43341 | 7194 | 0.166 | 0.5601 |
| chrUn_gl000244 | 39929 | 298 | 0.0075 | 0.1105 |
| chrUn_gl000245 | 36651 | 602 | 0.0164 | 0.1728 |
| chrUn_gl000246 | 38154 | 2999 | 0.0786 | 0.3505 |
| chrUn_gl000247 | 36422 | 2397 | 0.0658 | 0.3467 |
| chrUn_gl000248 | 39786 | 2998 | 0.0754 | 0.4061 |
| chrUn_gl000249 | 38502 | 0 | 0 | 0 |