Input data and parameters
QualiMap command line
| qualimap bamqc -bam /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_06_bamsort/case2.pair1_sorted.bam -nw 400 -hm 3 |
Alignment
| Command line: | "bismark -q --phred33-quals --bowtie2 --un --multicore 12 --output_dir /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_04_bismark --temp_dir /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_04_bismark --genome_folder /home/zhangwei/Genome/hg19_bismark -1 /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_03_adapterremoval/case2.pair1.truncated.gz -2 /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_03_adapterremoval/case2.pair2.truncated.gz" |
| Draw chromosome limits: | no |
| Analyze overlapping paired-end reads: | no |
| Program: | Bismark (v0.23.0) |
| Analysis date: | Tue Jan 12 17:43:29 CST 2021 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | /opt/tsinghua/zhangwei/Pipeline_test/o_WGBS-PE/intermediate_result/step_06_bamsort/case2.pair1_sorted.bam |
Summary
Globals
| Reference size | 3,137,161,264 |
| Number of reads | 5,105,250 |
| Mapped reads | 5,105,250 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 5,105,250 / 100% |
| Mapped reads, first in pair | 2,552,625 / 50% |
| Mapped reads, second in pair | 2,552,625 / 50% |
| Mapped reads, both in pair | 5,105,250 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Read min/max/mean length | 20 / 150 / 149.14 |
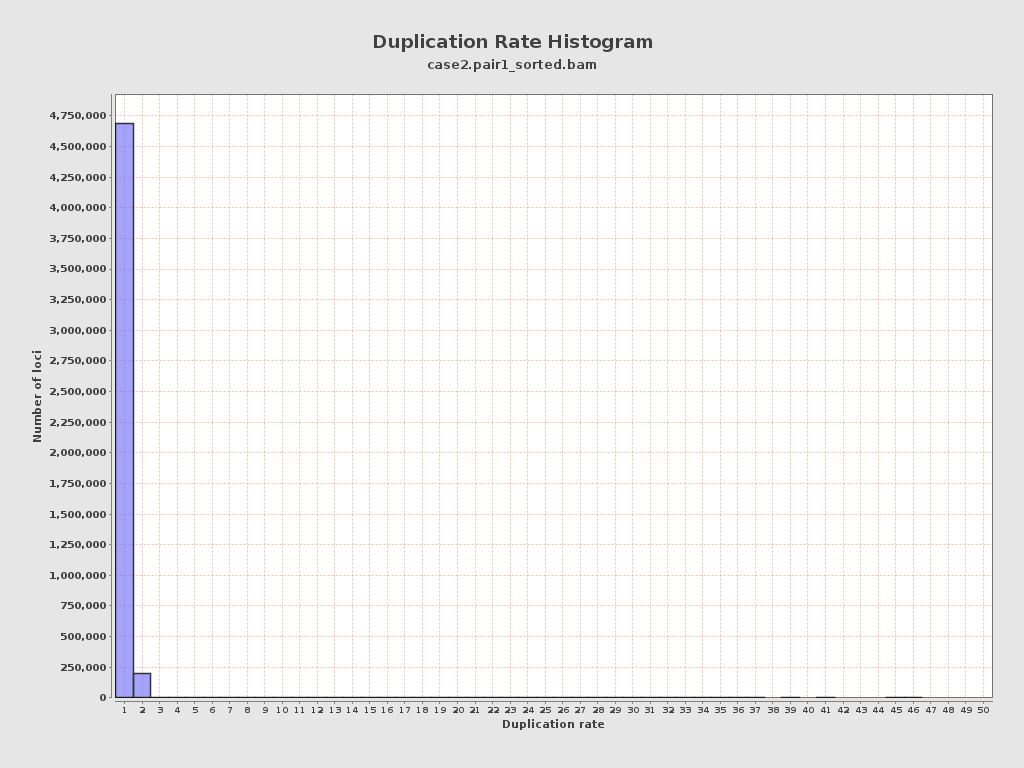
| Duplicated reads (estimated) | 214,402 / 4.2% |
| Duplication rate | 4.17% |
| Clipped reads | 0 / 0% |
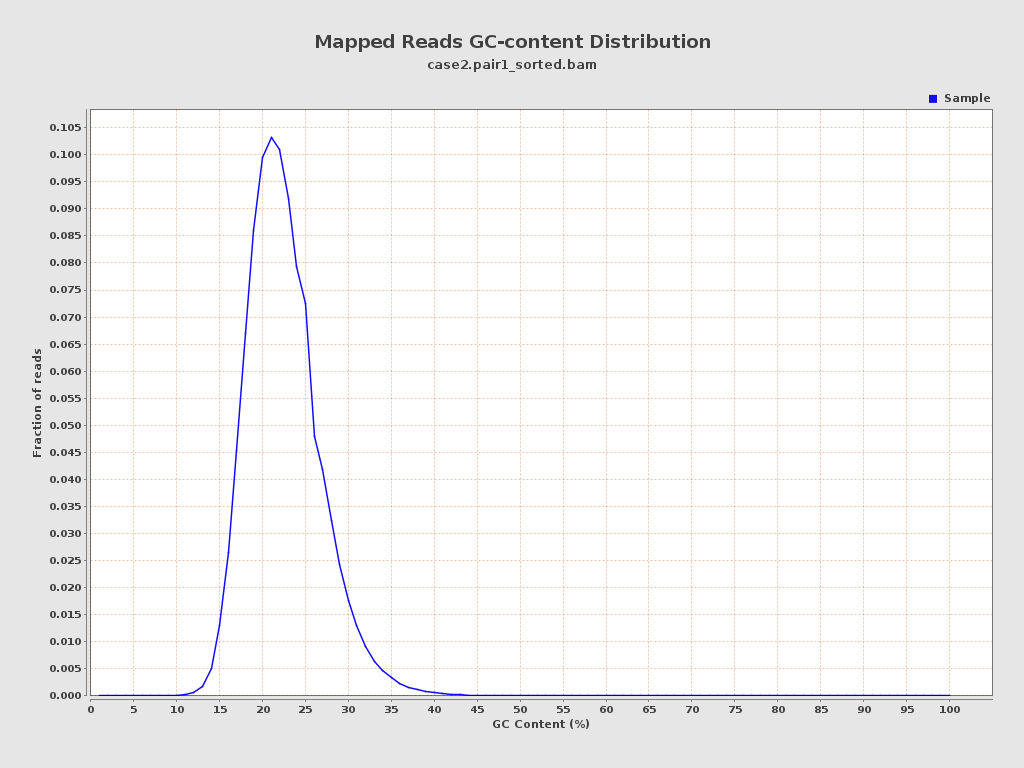
ACGT Content
| Number/percentage of A's | 296,076,478 / 39.02% |
| Number/percentage of C's | 83,505,437 / 11% |
| Number/percentage of T's | 296,160,353 / 39.03% |
| Number/percentage of G's | 83,116,949 / 10.95% |
| Number/percentage of N's | 91,233 / 0.01% |
| GC Percentage | 21.96% |
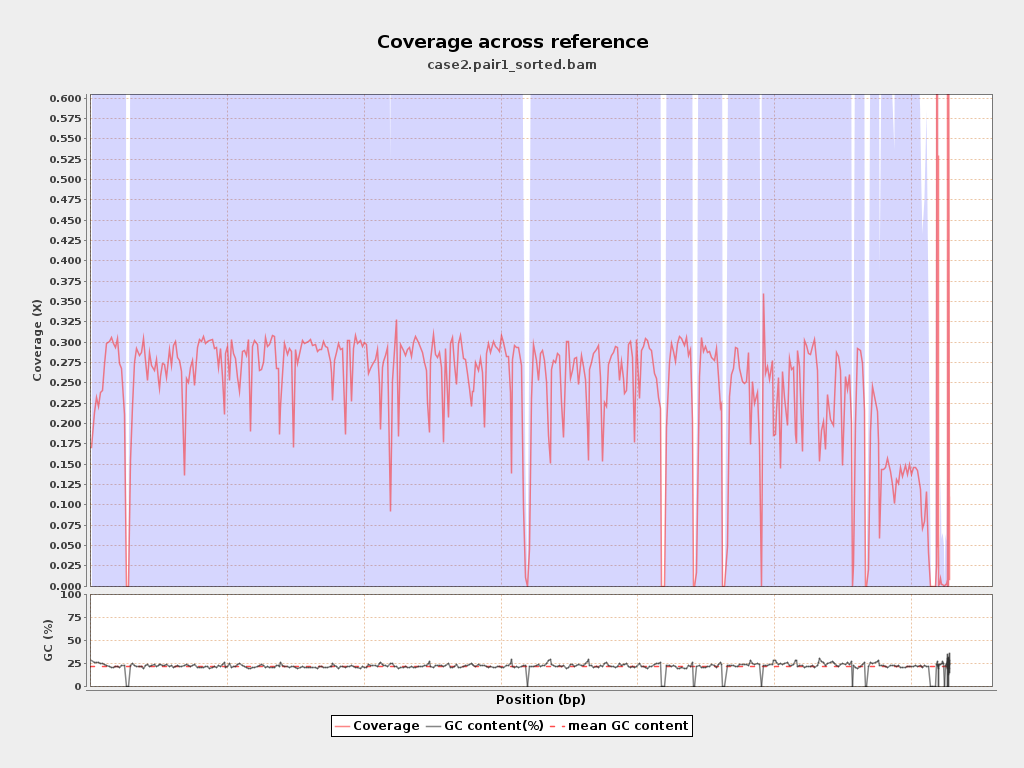
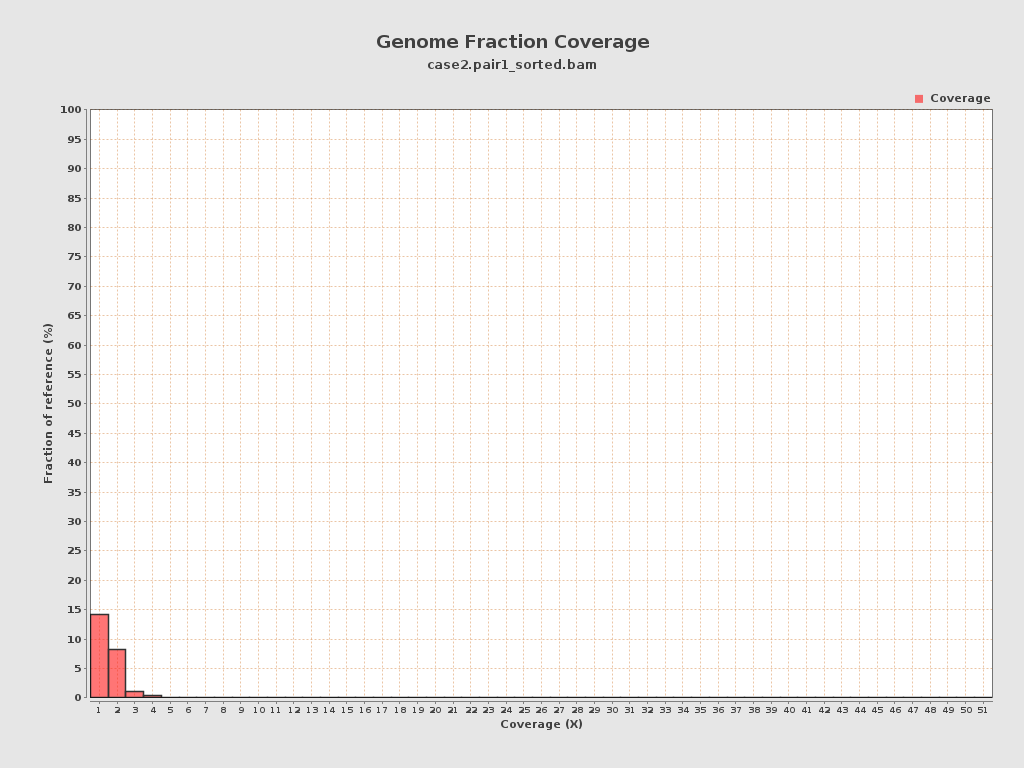
Coverage
| Mean | 0.2423 |
| Standard Deviation | 0.9439 |
Mapping Quality
| Mean Mapping Quality | 23.59 |
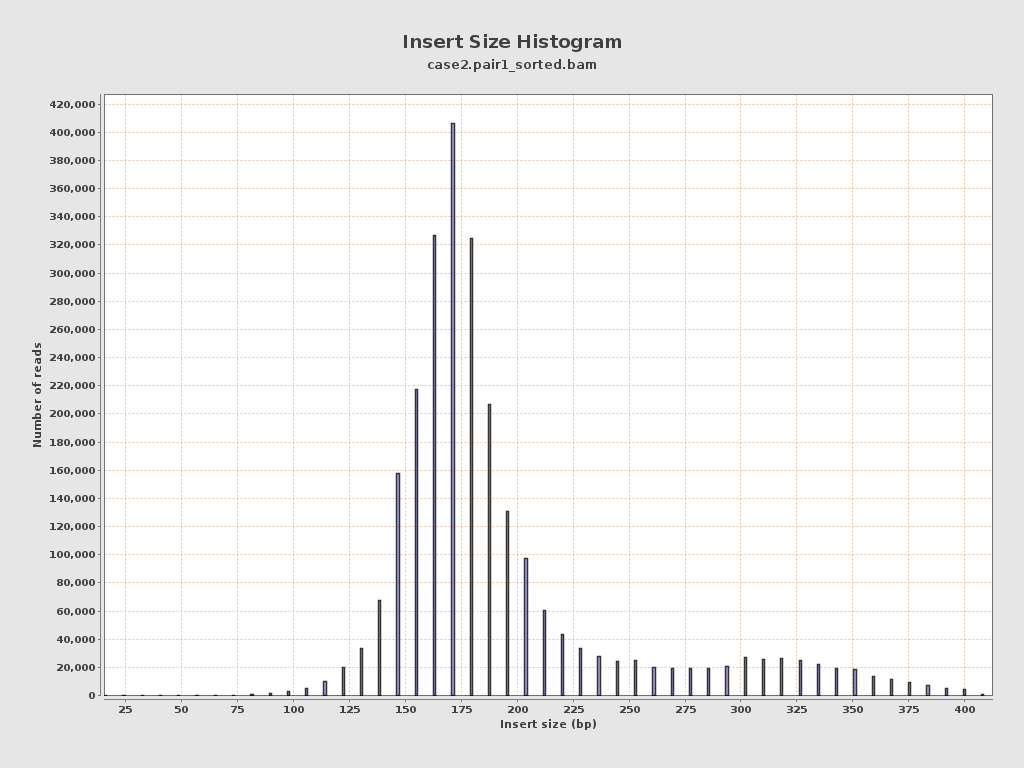
Insert size
| Mean | 198.61 |
| Standard Deviation | 57.36 |
| P25/Median/P75 | 167 / 180 / 204 |
Mismatches and indels
| General error rate | 19.68% |
| Mismatches | 147,109,330 |
| Insertions | 743,996 |
| Mapped reads with at least one insertion | 14.51% |
| Deletions | 442,194 |
| Mapped reads with at least one deletion | 8.32% |
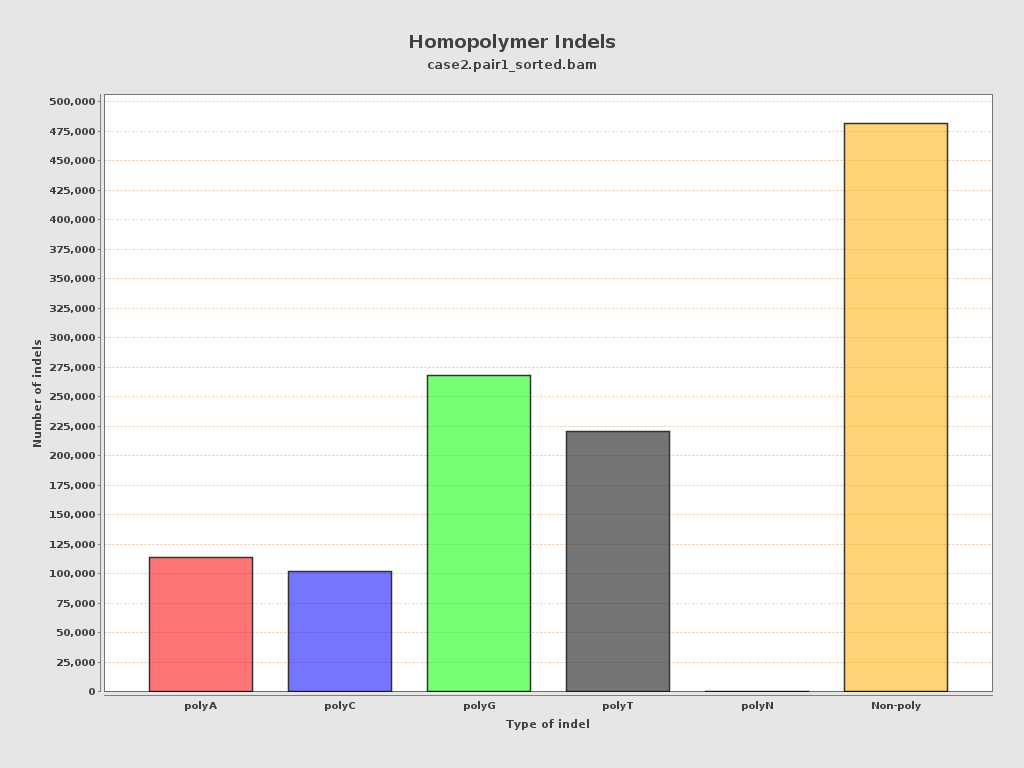
| Homopolymer indels | 59.39% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| chr1 | 249250621 | 61093747 | 0.2451 | 1.7199 |
| chr2 | 243199373 | 66783452 | 0.2746 | 0.7159 |
| chr3 | 198022430 | 55578115 | 0.2807 | 0.6996 |
| chr4 | 191154276 | 54161013 | 0.2833 | 0.7043 |
| chr5 | 180915260 | 50215841 | 0.2776 | 0.6982 |
| chr6 | 171115067 | 46837526 | 0.2737 | 0.8487 |
| chr7 | 159138663 | 42687102 | 0.2682 | 1.8202 |
| chr8 | 146364022 | 40079652 | 0.2738 | 0.7358 |
| chr9 | 141213431 | 30606444 | 0.2167 | 0.6249 |
| chr10 | 135534747 | 35857045 | 0.2646 | 1.155 |
| chr11 | 135006516 | 35256521 | 0.2611 | 0.6762 |
| chr12 | 133851895 | 36002152 | 0.269 | 0.686 |
| chr13 | 115169878 | 27608771 | 0.2397 | 0.6533 |
| chr14 | 107349540 | 24370420 | 0.227 | 0.638 |
| chr15 | 102531392 | 21686845 | 0.2115 | 0.6181 |
| chr16 | 90354753 | 20271023 | 0.2243 | 1.1292 |
| chr17 | 81195210 | 18642664 | 0.2296 | 0.6398 |
| chr18 | 78077248 | 21452223 | 0.2748 | 0.6949 |
| chr19 | 59128983 | 11503775 | 0.1946 | 0.6893 |
| chr20 | 63025520 | 15384207 | 0.2441 | 0.6539 |
| chr21 | 48129895 | 9838490 | 0.2044 | 0.6171 |
| chr22 | 51304566 | 7528211 | 0.1467 | 0.5139 |
| chrX | 155270560 | 20943808 | 0.1349 | 0.5198 |
| chrY | 59373566 | 2492592 | 0.042 | 0.3929 |
| chrM | 16571 | 72837 | 4.3954 | 2.3182 |
| chr1_gl000191_random | 106433 | 5090 | 0.0478 | 0.2929 |
| chr1_gl000192_random | 547496 | 68355 | 0.1249 | 0.4751 |
| chr4_ctg9_hap1 | 590426 | 7992 | 0.0135 | 0.161 |
| chr4_gl000193_random | 189789 | 100544 | 0.5298 | 1.0222 |
| chr4_gl000194_random | 191469 | 53850 | 0.2812 | 0.7131 |
| chr6_apd_hap1 | 4622290 | 1800 | 0.0004 | 0.0278 |
| chr6_cox_hap2 | 4795371 | 18386 | 0.0038 | 0.0808 |
| chr6_dbb_hap3 | 4610396 | 8368 | 0.0018 | 0.0561 |
| chr6_mann_hap4 | 4683263 | 10473 | 0.0022 | 0.0613 |
| chr6_mcf_hap5 | 4833398 | 6849 | 0.0014 | 0.0478 |
| chr6_qbl_hap6 | 4611984 | 8688 | 0.0019 | 0.0578 |
| chr6_ssto_hap7 | 4928567 | 15481 | 0.0031 | 0.0759 |
| chr7_gl000195_random | 182896 | 127366 | 0.6964 | 1.2305 |
| chr8_gl000196_random | 38914 | 1200 | 0.0308 | 0.2307 |
| chr8_gl000197_random | 37175 | 302 | 0.0081 | 0.0895 |
| chr9_gl000198_random | 90085 | 30464 | 0.3382 | 0.9629 |
| chr9_gl000199_random | 169874 | 264718 | 1.5583 | 4.8811 |
| chr9_gl000200_random | 187035 | 0 | 0 | 0 |
| chr9_gl000201_random | 36148 | 0 | 0 | 0 |
| chr11_gl000202_random | 40103 | 2995 | 0.0747 | 0.3441 |
| chr17_ctg5_hap1 | 1680828 | 10787 | 0.0064 | 0.1099 |
| chr17_gl000203_random | 37498 | 4469 | 0.1192 | 0.5068 |
| chr17_gl000204_random | 81310 | 8653 | 0.1064 | 0.4359 |
| chr17_gl000205_random | 174588 | 103397 | 0.5922 | 1.1472 |
| chr17_gl000206_random | 41001 | 1198 | 0.0292 | 0.2133 |
| chr18_gl000207_random | 4262 | 891 | 0.2091 | 0.5798 |
| chr19_gl000208_random | 92689 | 63835 | 0.6887 | 1.5372 |
| chr19_gl000209_random | 159169 | 2397 | 0.0151 | 0.1467 |
| chr21_gl000210_random | 27682 | 0 | 0 | 0 |
| chrUn_gl000211 | 166566 | 48961 | 0.2939 | 0.7182 |
| chrUn_gl000212 | 186858 | 49403 | 0.2644 | 0.7052 |
| chrUn_gl000213 | 164239 | 5049 | 0.0307 | 0.2231 |
| chrUn_gl000214 | 137718 | 128148 | 0.9305 | 1.566 |
| chrUn_gl000215 | 172545 | 2699 | 0.0156 | 0.1749 |
| chrUn_gl000216 | 172294 | 28016 | 0.1626 | 0.567 |
| chrUn_gl000217 | 172149 | 16456 | 0.0956 | 0.4006 |
| chrUn_gl000218 | 161147 | 91195 | 0.5659 | 1.014 |
| chrUn_gl000219 | 179198 | 88392 | 0.4933 | 0.9392 |
| chrUn_gl000220 | 161802 | 1112967 | 6.8786 | 18.1671 |
| chrUn_gl000221 | 155397 | 19432 | 0.125 | 0.5045 |
| chrUn_gl000222 | 186861 | 12464 | 0.0667 | 0.3302 |
| chrUn_gl000223 | 180455 | 1491 | 0.0083 | 0.1138 |
| chrUn_gl000224 | 179693 | 151768 | 0.8446 | 2.0539 |
| chrUn_gl000225 | 211173 | 43190 | 0.2045 | 0.6718 |
| chrUn_gl000226 | 15008 | 209046 | 13.929 | 41.2561 |
| chrUn_gl000227 | 128374 | 3007 | 0.0234 | 0.1956 |
| chrUn_gl000228 | 129120 | 6285 | 0.0487 | 0.3418 |
| chrUn_gl000229 | 19913 | 9238 | 0.4639 | 1.1162 |
| chrUn_gl000230 | 43691 | 10370 | 0.2373 | 0.6925 |
| chrUn_gl000231 | 27386 | 9825 | 0.3588 | 0.8998 |
| chrUn_gl000232 | 40652 | 23241 | 0.5717 | 1.048 |
| chrUn_gl000233 | 45941 | 9584 | 0.2086 | 0.6921 |
| chrUn_gl000234 | 40531 | 22071 | 0.5445 | 0.9736 |
| chrUn_gl000235 | 34474 | 22146 | 0.6424 | 1.1116 |
| chrUn_gl000236 | 41934 | 298 | 0.0071 | 0.1126 |
| chrUn_gl000237 | 45867 | 8698 | 0.1896 | 0.6248 |
| chrUn_gl000238 | 39939 | 598 | 0.015 | 0.1642 |
| chrUn_gl000239 | 33824 | 1457 | 0.0431 | 0.3234 |
| chrUn_gl000240 | 41933 | 10966 | 0.2615 | 0.7015 |
| chrUn_gl000241 | 42152 | 36339 | 0.8621 | 1.3039 |
| chrUn_gl000242 | 43523 | 1206 | 0.0277 | 0.213 |
| chrUn_gl000243 | 43341 | 4790 | 0.1105 | 0.4277 |
| chrUn_gl000244 | 39929 | 604 | 0.0151 | 0.1629 |
| chrUn_gl000245 | 36651 | 896 | 0.0244 | 0.2243 |
| chrUn_gl000246 | 38154 | 3577 | 0.0938 | 0.4585 |
| chrUn_gl000247 | 36422 | 1726 | 0.0474 | 0.2901 |
| chrUn_gl000248 | 39786 | 1500 | 0.0377 | 0.2548 |
| chrUn_gl000249 | 38502 | 300 | 0.0078 | 0.1184 |